We selected 3 very different biomedical problems to use as test beds for our algorithms and to drive the development of new algorithms that meet the needs of biomedical researchers.
Cancer
The Cancer Signaling Pathways project seeks to discover the genomic drivers of tumors and the cell signaling pathways that are being abnormally affected, driving the development of cancer. The ability to discover and model these causal relationships accurately is a key step in more fully realizing precision cancer diagnosis, prognosis, and therapy.
We are analyzing The Cancer Genome Atlas (TCGA) data for 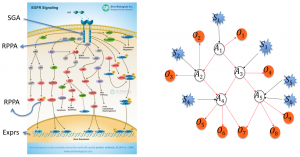 breast and head and neck cancer to discover which somatic genetic alterations are causal drivers of a given tumor and which are causal drivers shared across different tumors as well as the causal relationships among signaling proteins in cancer pathways.
breast and head and neck cancer to discover which somatic genetic alterations are causal drivers of a given tumor and which are causal drivers shared across different tumors as well as the causal relationships among signaling proteins in cancer pathways.
Available data include measurements of somatic mutations, copy number alterations, DNA methylation, gene expression, microRNA expression, and reverse phase protein array (RPPA) data, which we collectively refer to as functional genomic data, as well as additional clinical data for many tumor samples submitted by the University of Pittsburgh to TCGA.
We will take advantage of the Pittsburgh Genome Resource Repository, a collaborative effort of the University of Pittsburgh, the Pittsburgh Supercomputing Center, and UPMC, for faster data access and analysis.
Lung Disease
Physicians depend on clinical tests for diagnosis, but these are often inconclusive at early stages of disease, and sometimes cannot distinguish between disease subcategories.
The Lung Diseases project aims to discover the cellular factors that lead to susceptibility and progression of chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis.
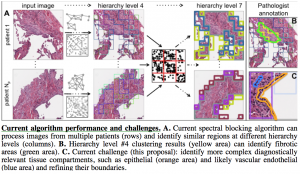 We are analyzing data from the Lung Genomics Research Consortium and the Lung Tissue Research Consortium to discover and model causal relationships between molecular variables, clinical variables, and image features to characterize disease mechanisms and predict disease severity. The data include high-resolution tissue images as well as SNP, DNA methylation, mRNA expression, and microRNA expression data from patient biospecimens.
We are analyzing data from the Lung Genomics Research Consortium and the Lung Tissue Research Consortium to discover and model causal relationships between molecular variables, clinical variables, and image features to characterize disease mechanisms and predict disease severity. The data include high-resolution tissue images as well as SNP, DNA methylation, mRNA expression, and microRNA expression data from patient biospecimens.
However, before we can use omics data in causal modeling, we must address an important issue: most clinical omics data are acquired from homogenized tissues with multiple cell types. Causal modeling of cellular function is facilitated when measured variables are derived from single cells or, at the very least, from homogeneous cell type populations. Thus, we will first partition the existing omics measurements into relevant tissue types by using the matched images for guidance (see figure for preliminary results).
First, we will identify tissue compartments using our Spectral Blocking approach, which computationally groups image patches into tissue compartments of shared cell type, and the guidance of our pathologist collaborator, Dr. Frank Schneider.
We will then partition the omics signals across patients (tissues) using one of several existing methods for deconvolution or unmixing of genomic signals from heterogeneous samples.
Finally, we will use causal discovery algorithms to identify molecular interactions, signatures, and pathways that are associated with each disease.
Brain
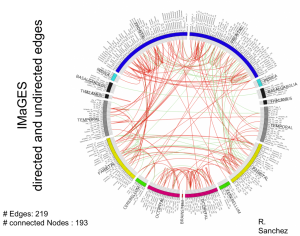 The Brain Functional Connectivity project seeks to discover the causal influences among small spatial regions of the human brain. The ability to identify accurately active causal pathways (“effective connections”) resolved at the voxel level for the entire cortex is within reach and offers the prospect of much finer diagnoses and classifications of disorders and improved monitoring of treatment effects.
The Brain Functional Connectivity project seeks to discover the causal influences among small spatial regions of the human brain. The ability to identify accurately active causal pathways (“effective connections”) resolved at the voxel level for the entire cortex is within reach and offers the prospect of much finer diagnoses and classifications of disorders and improved monitoring of treatment effects.
We use fMRI data that can record the activity of approximately 2mm3 regions (voxels) of the brain about every two seconds. These regions define thousands of variables that we analyze to generate a causal network of functional influence.
We are performing this analysis on fMRI data from individuals with autism spectrum disorder and individuals who are neurotypicals. We want to characterize causal-network differences between these groups, as well as differences among those individuals with autism spectrum disorder. We plan to perform a similar investigation on fMRI data of individuals with schizophrenia.
